Super-Enhancers
A repertoire of super-enhancers
A core collection of super-enhancers with their unique charactheristics
Introduction
This repertoire contains a set of super-enhancers along with typical enhancers and stretch enhancers on selected cell-lines / tissues. In addition, a series of statistical comparisons were carried out between super-enhancers and typical/stretch enhancers. These analyses demonstrate that super-enhancers are a unique subclass of enhancers, with distinct characteristics from typical enhancers and, more importantly, stretch enhancers.
Abbreviations: Super-Enhancer, SE; Typical Enhancer, TE; Stretch Enhancer, StrE
Data access
Super-enhancers and typical enhancers identified based on ChIP-seq data from the ENCODE project
We collected the data and provide downloadings (hg19 assembly) below:
- Super-enhancers [SE Download]
- Typical enhancers [TE Download]
Note: Above are links for bulk downloadings, please check [here] for an additional table to download the data of individual cell types.
Super-enhancers, stretch enhancers, and typical enhancers in the eight extensively studies cell lines
We collected the data and provide downloadings (hg19 assembly) below:
- Super-enhancers [SE Download]
- Stretch enhancers [StrE Download]
- Typical enhancers [TE Download]
Note: Above are links for bulk downloadings, please check [here] for an additional table to download the data of individual cell types.
Comparison to typical enhancers
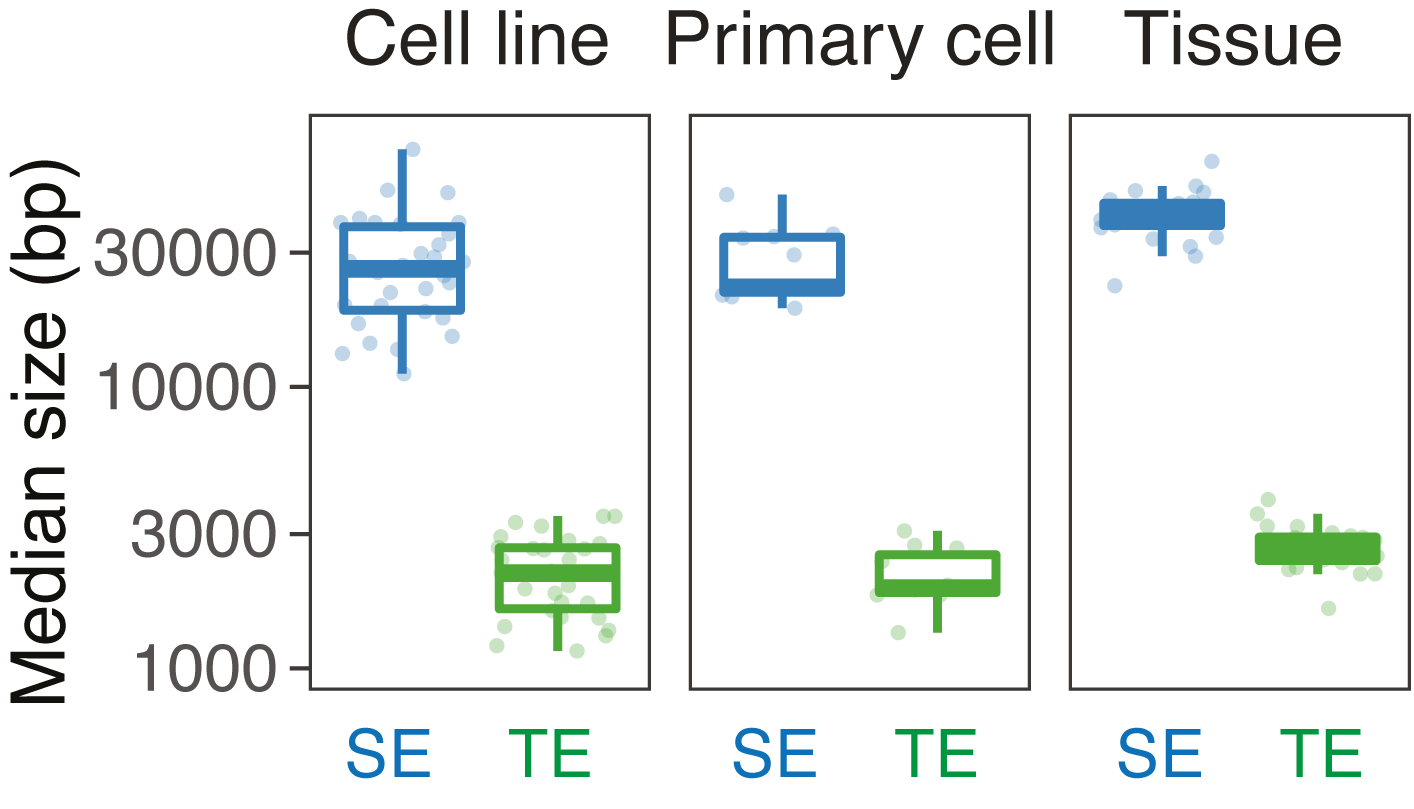
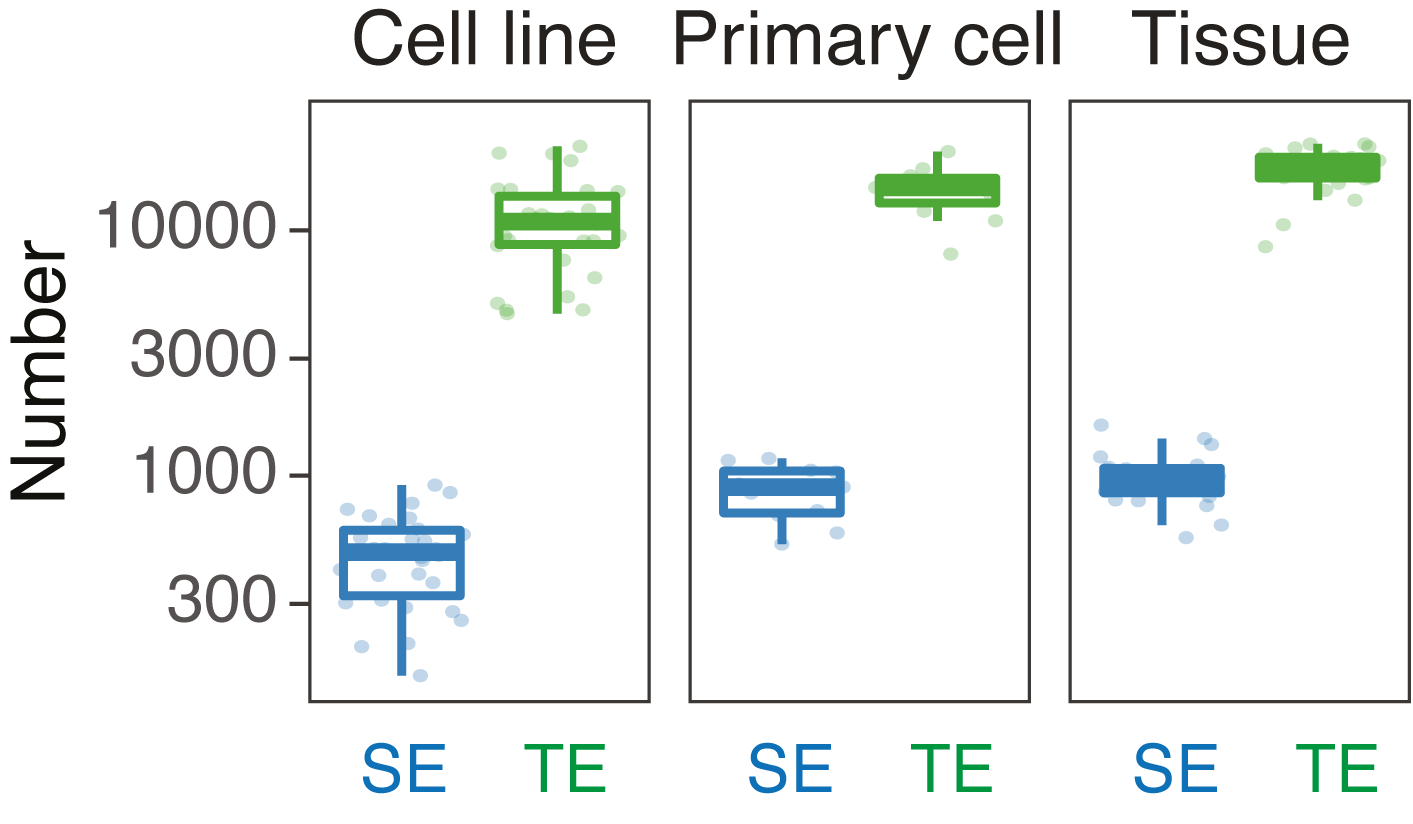
We compared the size and the number of SEs and typical enhancers (TE) in each cell type.
SE vs TE: median size
SE vs TE: numbers
Please click [here] to recap the analysis.
Comparison to stretch enhancers
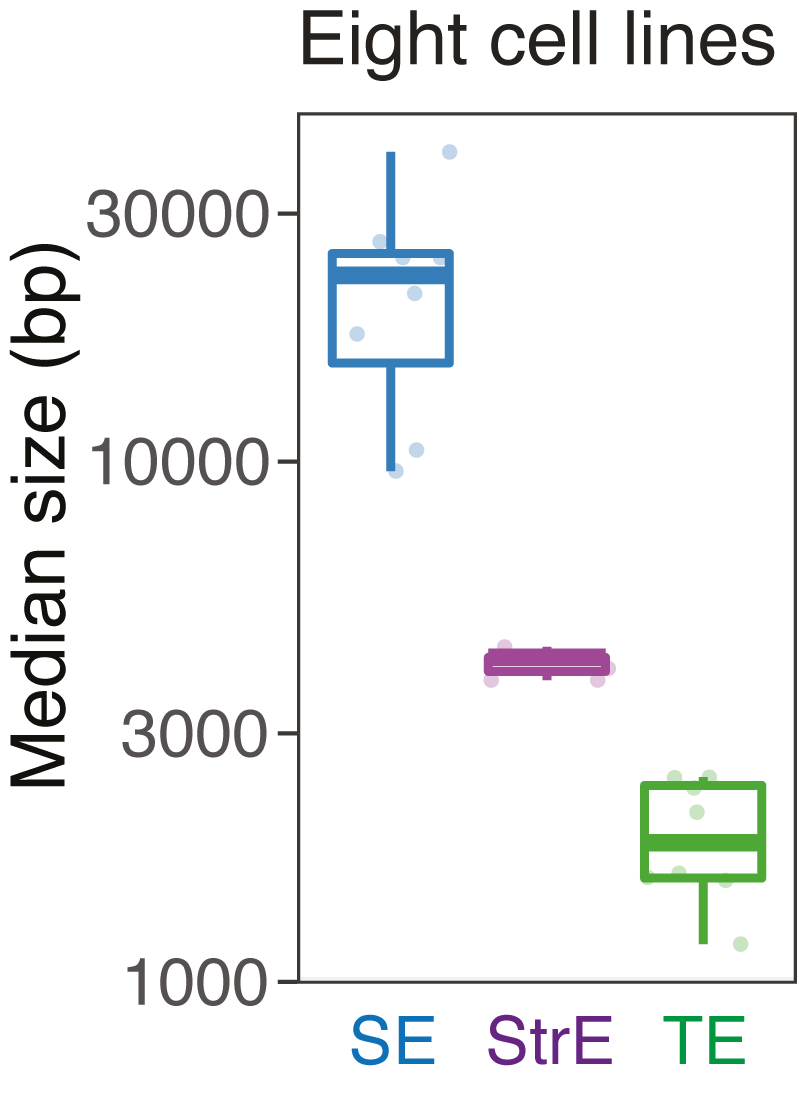
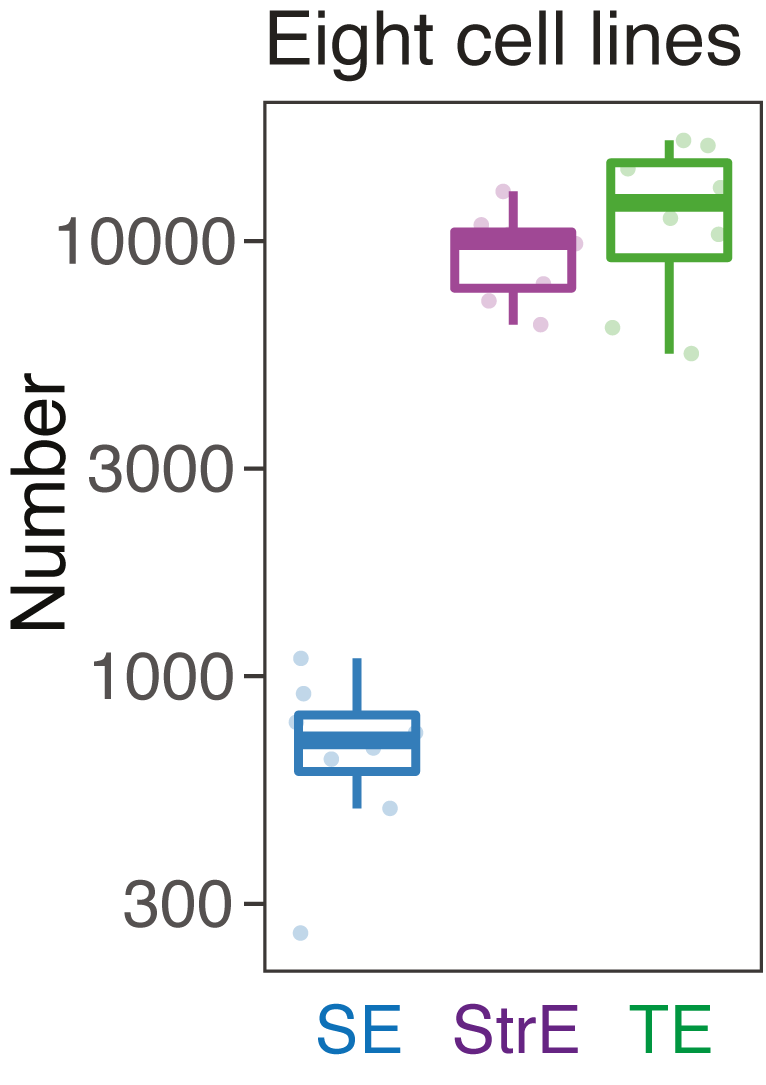
We compared the size and the number of SEs, stretch enhancers (StrE) and typical enhancers (TE) in each cell type.
SE vs StrE vs TE: median size
SE vs StrE vs TE: numbers
Please click [here] to recap the analysis.
Super-enhancer landscape, hierarchical clustering, and principal component analysis
Landscape on chromosomes
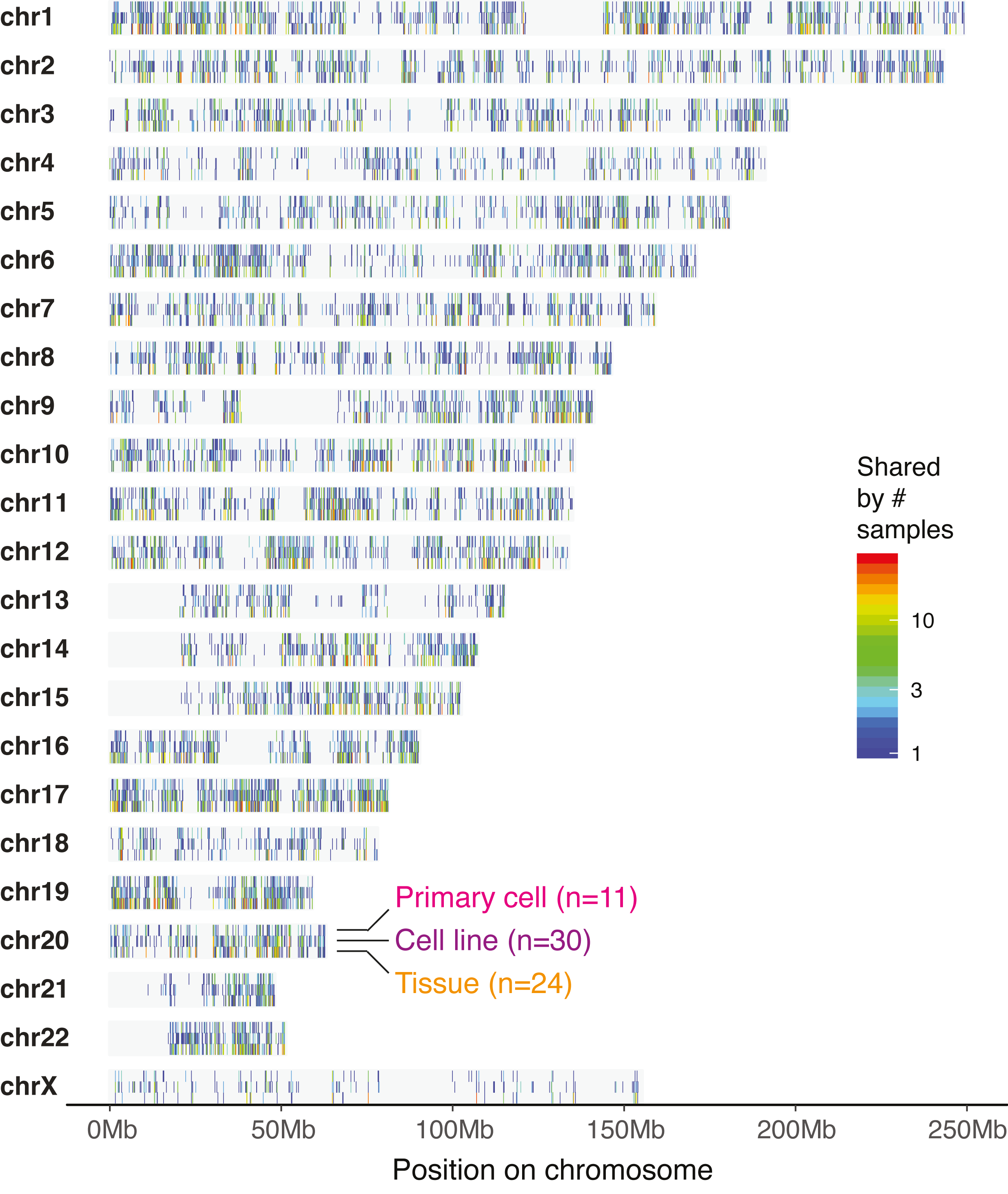
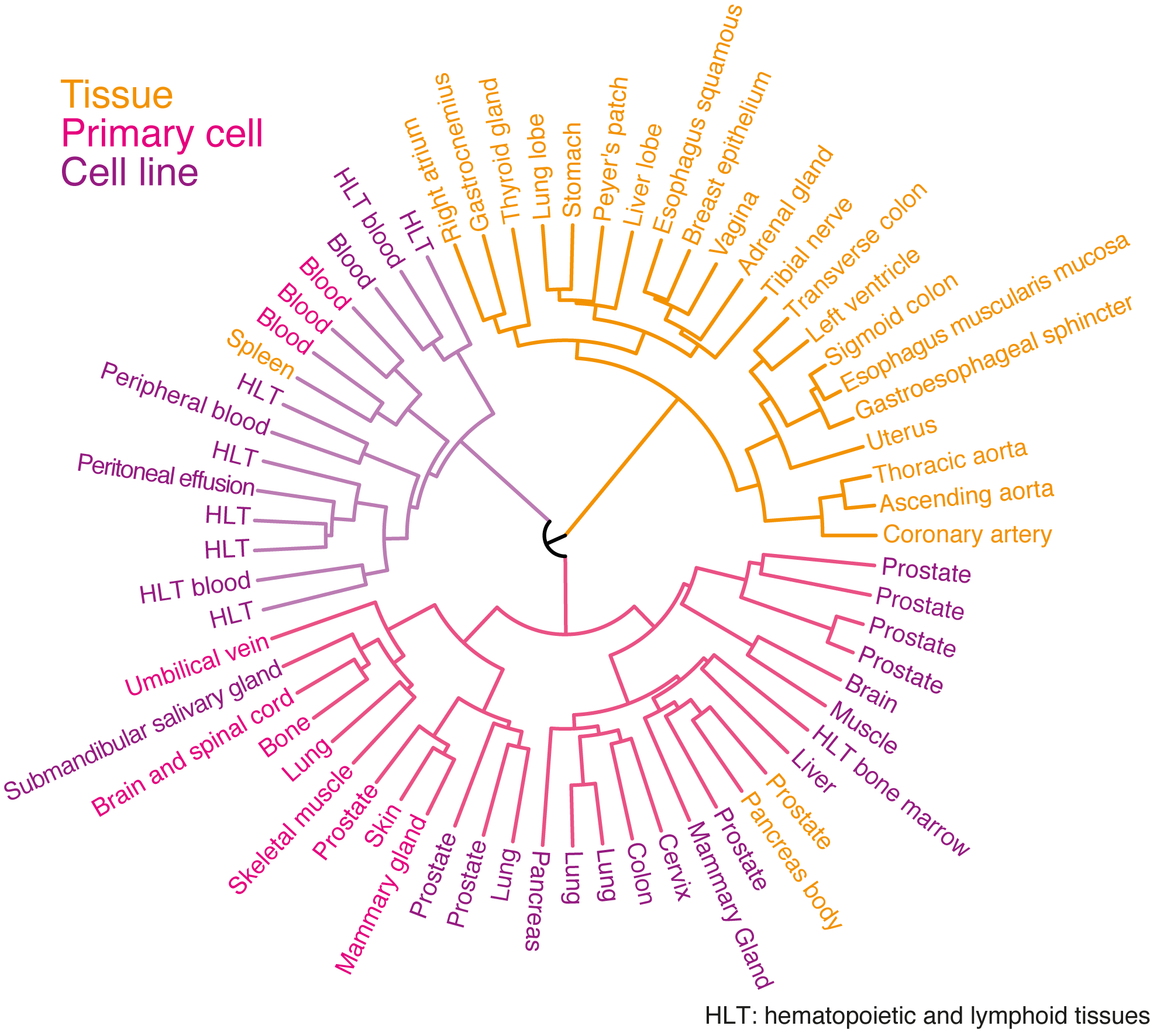
Hierarchical clustering
Hierarchical clustering based on SE cooccurrence Jaccard distance between paris of samples
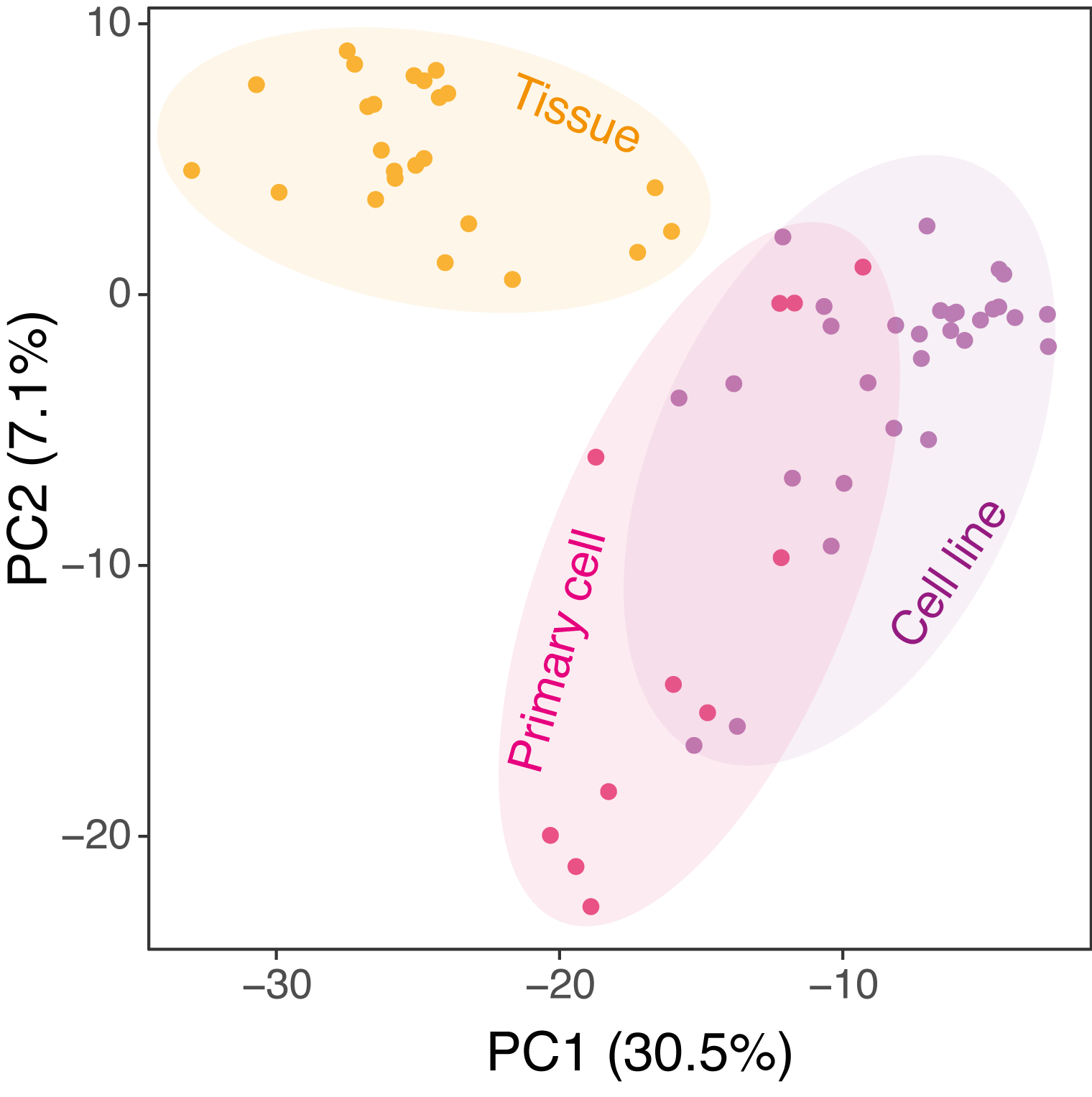
Principal component analysis
Principal component analysis based on SE occurrence matrix on all the samples
Please click [here] to recap the analysis.
Support or Contact
Xi Wang [xiwang (at) njmu (dot) edu (dot) cn] & Jian Yan [jian (dot) yan (at) cityu (dot) edu (dot) hk]
Citation
Xi Wang, Murray J. Cairns, Jian Yan. Super-enhancers in transcriptional regulation and genome organization. Nucleic Acids Research, 2019, 47(22): 11481-11496.